Offline tool
An offline tool for identification of targets from pathogen proteomes, protein-protein interaction network and genome scale metabolic model.
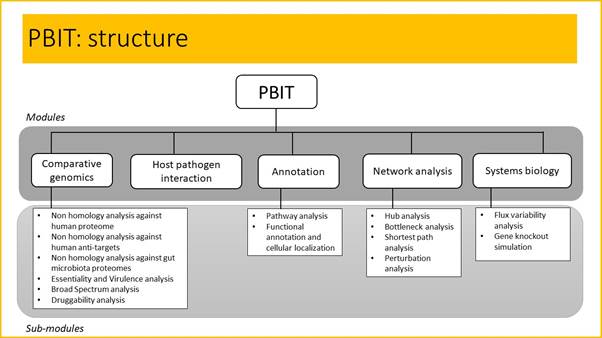
Offline PBIT contains 5 modules and respective submodules:
- Comparative genomics – This module is based on sequence comparison method of pathogen proteins with the human proteome, anti-targets, gut microbiota, essential and virulent proteins and known drug targets, to identify ideal targets. The functionality of each module remains same as that of online PBIT. This module can also be plugged in with the annotation module.
Submodules:
- Non homology analysis against human proteome
- Non homology analysis against human anti-targets
- Non homology analysis against gut microbiota proteomes
- Essentiality and virulence analysis
- Broad spectrum analysis
- Druggability analysis
These submodules can function independently or combined into a pipeline for target identification.
- Host Pathogen Interaction - This module identifies targets that share sequence similarity to microbial proteins known to interact with human proteome, human anti-targets, human gut microbiota.
- Annotation – This module annotates pathway and functionality of the query protein sequences from UniProt KB database.
Submodules:
- Pathway analysis
- Functional annotation and subcellular localization
- Network Analysis – This module calculates network topological features.
Submodules:
- Hub analysis
- Bottleneck analysis
- Shortest path analysis
- Perturbation analysis
- Systems biology – This module determines essentiality of reactions and genes using flux variability analysis and single gene knockout.
Submodules:
- Flux variability analysis
- Gene knockout simulation
Modules |
Input |
Parameters |
Output |
Comparative genomics |
FASTA formatted protein sequences |
Choice of submodules
Percent identity
Alignment length cut off
E-value |
FASTA formatted protein sequences |
Host Pathogen Interaction |
FASTA formatted protein sequences |
Percent identity
E-value |
FASTA formatted protein sequences |
Annotation |
FASTA formatted protein sequences |
Choice of submodules |
Annotated protein identifiers in excel format |
Network Analysis |
Edge list in CSV format |
Choice of submodules |
Text file containing output of selected submodules |
Systems biology |
- Metabolic model in JSON format
- Gene list from model (optional)
|
Choice of submodules
Reaction ids & bounds (optional) |
Excel file containing output of selected submodules |
Command line interface for offline PBIT
From Windows command line:
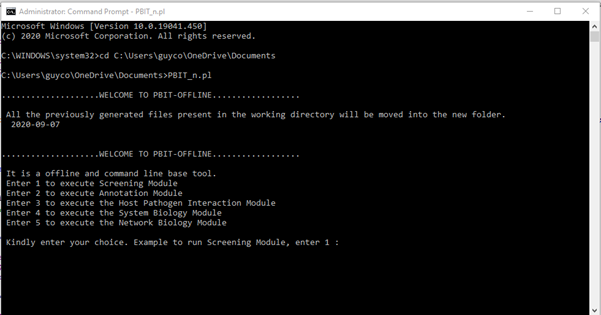
For detailed information on installation and dependencies, please download the manual for PBIT
Further reading:
- Orth, Jeffrey D., Ines Thiele, B. Ø. P. What is flux balance? Nat. Biotechnol. 28, 245–248 (2010).
|
|
Citation:
Mukherjee S, Kundu I, Askari M, Barai R S, Venkatesh K V, Idicula-Thomas S. Exploring the druggable proteome of Candida species through comprehensive computational analysis. Genomics, Volume 113, Issue 2,2021, Pages 728-739. (https://doi.org/10.1016/j.ygeno.2020.12.040.)
|

